R package for modelling microbial Populations and Communities
Microbial communities perform highly dynamic and complex ecosystem functions that impact plants, animals and humans. MicroPop is a piece of software which attempts to model the dynamics of microbial interactions for a wide range of microbial systems. It is the result of an interdisciplinary project resulting from a collaboration between Helen Kettle and The Rowett Institute (Petra Louis and Harry Flint).
microPop Tutorial
If you want an introduction on how to get started with microPop please download the zip file below. Once you have extracted the files you will see the following folders: Html, Inputfiles, RCode, Rmarkdown and a Readme.txt file. Click on the readme file to begin. I have also added the two html files below which you can simply read if you don't want to get involved with running the code.
Tutorial web page on getting started with microPop.
Tutorial web page with exercises solutions.
ZipFile for Tutorial on getting started with microPop.
Plotting microPop networks using visNet
Plotting networks using the shiny app
What does microPop do?
For a full description of microPop's capability see Kettle et al 2018 (and the supporting information); a brief overview is given below.
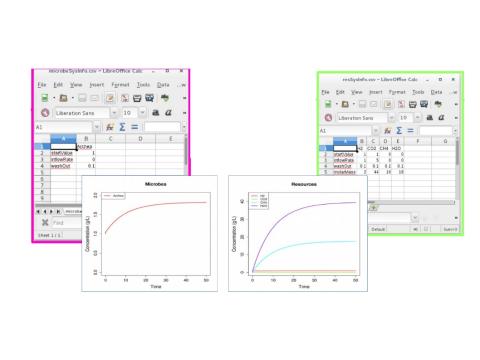
- microPop simulates the dynamics and interactions of microbial populations by solving a system of ordinary differential equations that are constructed automatically based on a description of the system (which can be specified via csv files).
- Data frames (R objects) for a number of microbial functional groups and default functions for rates of microbial growth, resource uptake, metabolite production are provided but can be modified or replaced by the user.
- micropop can simulate growth in a single compartment (e.g. bio-reactor) or 'compartments' in series (e.g. human colon) or in a simple 1-d application (e.g. phytoplankton in a water column).
- A microbial functional group may contain multiple strains in order to study adaptation and diversity or parameter uncertainty.
- Simple interactions between viruses (bacteriophages) and bacteria can also be included in micropop.
- I have also developed a R Shiny App to visualize the microbial networks contained in microPop.
What is it?
MicroPop is a piece of open source software written in R (a free software environment that runs on Linux, Windows and MacOS). It is hosted here on CRAN.
How do I install it?
To use microPop simply download R and when in R type install.packages('microPop').
Testimonials for microPop
"This work is an outstanding effort that enables a hands-on approach for linking compositional data to functional predictions. It allows direct, hypotheses-driven simulations and functional data interpretation. The user-friendly implementation makes it also accessible to wet-labs that are otherwise not specialized on microbiome modelling. The flexible framework can be used for a variety of microbial systems and environments and promises access to functional-dynamical interpretation of data and processes as well generation of predictions and new hypotheses. The automated generation of an ODE system that is fully accessible to user-defined system properties clearly fills a method gap in microbial ecology." - - anonymous reviewer 1.
"This package adds a new toolset for reproducible research in microbial population modelling. Its novelty lies not just in the fact that its main form of input are system descriptions, but also because it gives the user the freedom to modify the models themselves." -- anonymous reviewer 2
"The microPop package developed by Kettle et al offers a powerful platform for studying microbial interactions via mathematical modelling. The software is relatively easy to operate and has the flexibility for tackling diverse scenarios." -- anonymous reviewer 3
