Rapid typing of E. coli isolates using whole cell MALDI mass spectrometry
Foodborne pathogens, including E. coli O157 and related strains (collectively called enterohaemorrhagic E. coli– EHEC), continue to cause public health problems, requiring typing of strains during outbreak investigations. Identification of bacterial strain by conventional means can be laborious and time-consuming. As an alternative, MALDI mass spectrometry (MS) proteomic profiles are quick and cost-effective to obtain. The possibility of using them for typing has been explored in collaboration with the Moredun Research Institute.
MS is a technologically advanced approach for detecting biological molecules. The complex spectra obtained from the spectrometer require a series of pre-processing steps to convert these data into formats suitable for bacterial identification and typing. High-quality pre-processing usually includes a combination of smoothing, baseline correction, peak extraction and alignment, and normalisation procedures. BioSS has produced a quick, widely applicable, semi-automatic and statistically robust MS data pre-processing pipeline to facilitate these tasks. Once processed, the data can be analysed using statistical clustering and phylogenetic methods to investigate relationships amongst strains. Relationships identified by MALDI-MS are also being used as input for a predictive model based on shrinkage linear discriminant analysis. Taken as a whole, this approach demonstrates greater than 90% accuracy and allows users to identify the most significant spectral features involved in distinguishing between bacterial strains.
Application of these methods and associated software to 92 EHEC isolates shows a good level of differentiation and reproducibility among biological samples. When interpreted using these new quantitative tools, MALDIMS is showing promise as a typing method that could be rapidly applied during an infection outbreak, supporting better public health epidemiology.
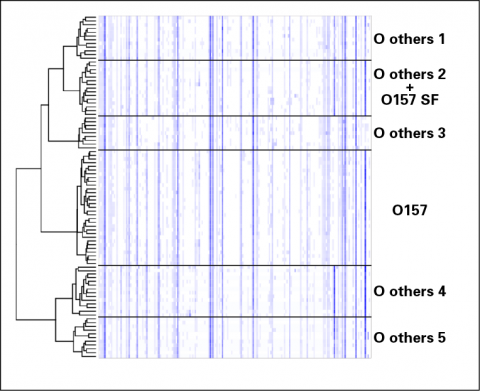
For the 92 E. coli – EHEC isolates, the dendrogram on the left-hand side represents the similarity relationships identified by statistical clustering based on their MS fingerprints. The E. coli O157 serogroup is neatly distinguished from other EHEC O serogroups, including sorbitol-fermenting (SF) O157. A heat map of the processed MS intensities across the 4.3 – 9.8 kDa m/z range (darker shading corresponds to higher concentrations) demonstrates the key differences between the isolates clustered into the identified groups.
This work was undertaken by Javier Palarea-Albaladejo and Frank Wright.

