Classifying tumour biopsies
Colorectal cancer is the fourth most common cause of death from cancer, accounting for 8% of all cancer deaths. It is of particular interest to nutritional scientists, as it is known to be strongly influenced by diet.
Scientists from the Rowett Institute of Nutrition and Health have developed a targeted in-house multiplex gene array to measure expression of 21 markers (genes) known to be involved in the development of colorectal cancer. This array has been used to study differences between normal, polyp and tumour colon biopsies (58 samples in total), which represent different stages of the disease. Analysing the differential gene expression patterns, we can find genes of diagnostic value for staging the diseases, as well as learn about the gene regulatory mechanisms involved.
BioSS applied orthogonal partial least squares discriminant analysis (OPLS-DA) as a method to visualize the main patterns in the data (see plot), as well as to identify classifiers for the three sample types. Leave-one-out cross validation showed that 100% of the normal samples, 88% of the polyps and 94% of the tumours could be classified correctly in this data set. [In particular, the study showed that epithelial genes such as NOX1 and LGR5 express at high levels in polyps, which might be helpful in predicting individuals with a propensity for tumour development.
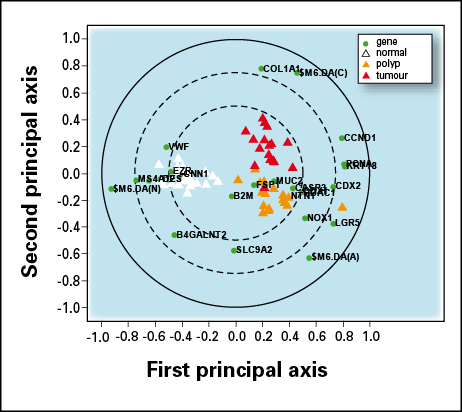
Bi-plot obtained from OPLS-DA. The first component (horizontal axis) separates normal (white triangles) from other samples, whereas the second component (vertical axis) distinguishes between polyp (yellow triangles) and tumour (red triangles) samples. Dark circles represent the genes, highlighting the wide transition between expression of genes such as $M6DA(N) in normal samples and the expression of a different set of genes such as CCND1 in unhealthy samples.

